New CBTN Data Available
Posted on

As part of Project Accelerate, CBTN is pleased to announce that the somatic mutations across over 1,800 newly characterized tumors are now available for immediate public access on PedcBioPortal! These have been incorporated into a provisional release as part of the Pediatric Brain Tumor Atlas, which now includes a total of over 3,055 samples with whole genome and RNA-seq.
What does “provisional” mean?
In the interest of rapid availability to the community, the provisional release consists of automated output from the Kids First DRC somatic pipelines without further downstream analysis and review. This data is pre-publication and available without embargo. As we continue to optimize your PedcBioPortal user experience, perform further checks and filtering, you can be sure that we will update the this powerful trove of data will remain available in place to continue to provide what we view as the best dataset currently available to the global research community.
Some important notes for this release:
- Gene annotation models have been updated from ENSEMBL v93/GENCODE 27 -> ENSEMBL v105/GENCODE39
- Many more fusion candidates are included due to updates in fusion calling software;, we anticipate many of these may be filtered out in the future with updated criteria
- The new samples have not undergone OpenPedCan processing so are missing subtyping and deeper analytic quality checks
- The new samples have been checked for tumor/normal mismatches
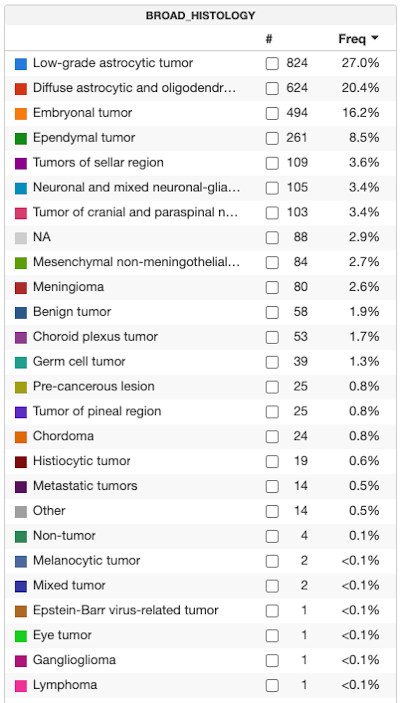
We anticipate the next major revision of this dataset to occur over the next two months, following OpenPedCan data processing.being done after the OpenPedCan processing over the next two months. An updated tally view of all the broad histologies now available is featured below. OneAn example of our what we anticipated being improvements will be with further analytic processing tois better classify any tumor types currently listed asication of the “NA” orand “Other” tumor types.
We’re excited for you to begin using these resources and look forward to your feedback as we continue to advance this resource.