Q&A with Dr. Chad Creighton - Exploring the Role of Somatic Structural Variation in Childhood Brain Cancer
Posted on
by
Jonathan Waller
The mechanisms that push healthy human cells to become cancerous are incredibly complicated and still not fully understood by science. What causes the genes within a cancer cell to copy, to fuse, or to express themselves in a given way, and how can this activity lead to the development of various forms of cancer, including pediatric brain tumors?
To explore complex questions like this, researchers require a LOT of genomic data, and powerful analysis tools to compare the terabytes of information needed make new and unexpected insights. The Children’s Brain Tumor Network’s Pediatric Brain Tumor Atlas is the world's largest collection of childhood brain tumor data and is available to access in real-time by researchers located all over the globe. The PBTA data contains more than 30 different subtypes of childhood brain tumors, representing more than 1,100 unique research subjects.
Using this valuable resource, scientists at institutions worldwide are able to launch new projects to study childhood brain cancer in ways never done before. We recently spoke with Dr. Chad Creighton, Professor of Medicine and Co-Director of Cancer Bioinformatics at Baylor College of Medicine, whose team utilized DNA and RNA data from hundreds of study participants within the PBTA for a major study on how Somatic Structural Variation within gene copying may drive the development of cancer cells within a child’s brain.
Below, he shares what his team discovered and how these new insights could help in the development of better precision medicine approaches for treating cancer in children.
Your team was conducting research into the role of Somatic Structural Variation (SSV) in pediatric brain cancer, specifically in how SSVs influence the way certain human genes behave. What is the potential in looking at how SSVs influence gene behavior, and how does that relate to how cancer cells form and multiply?
Cancer arises from DNA damage affecting the structure and expression of specific genes. "Oncogenes" are like the accelerator in a car. They drive cell proliferation, by virtue of these genes being hyperactive or over-expressed. In contrast, "Tumor suppressor" genes are like the brakes on a car. These genes would normally stop the cancer cells from proliferating. Cancers prefer DNA alterations that either keep oncogenes active or delete or inactivate tumor suppressor genes.
Until recently, scientists have focused on DNA alterations within the actual genes. But more recently, alterations in non-coding DNA regions have been found to affect the regulation of nearby genes. In our study, we focused on a class of DNA alterations called somatic structural variants (SSVs). SSVs involve a piece of DNA from one part of the genome being rearranged to join with a piece of DNA from a different part of the genome. The point where the two DNA pieces join is called a "breakpoint". When this happens, the normal regulation of genes next to the SSV DNA breakpoint is often changed. The rearrangement may involve something called an "enhancer" being placed next to the gene, which causes the gene to be over-expressed. Genes with oncogene roles may be over-expressed due to several different mechanisms involving SSVs.
In our study of pediatric brain tumors, we examined SSVs in combination with gene expression patterns. We could identify hundreds of genes that were over-expressed in association with a nearby SSV breakpoint, many of which are known oncogenes. In some instances, SSVs had breakpoints within a gene, leading to what is known as gene fusions, a new gene being formed by parts of two different genes being joined together. In other instances, the SSV breakpoint happened nearby the gene, and while the gene sequence itself was not altered, the regulation of the gene was impacted, and the gene was turned on when it should have been turned off.
This type of analysis isn’t new - somatic structural variation has been studied in adult cancers before. What promising findings have been uncovered in earlier research efforts?
In adult cancers, the phenomenon described above has been found by our group and others to impact hundreds of specific genes in different cancer types. As compared to adult cancers, pediatric tumors may involve a different set of oncogenes and tumor suppressors.
Your team’s research was just published in a new paper at Nature Communications, where you highlight the fact that somatic structural variation has never before been analyzed at such a large scale, including a broad cross-section of pediatric cancers. Why hasn’t large-scale SSV analysis across children and adults been done before?
Large-scale SSV analysis has been done just recently in adult cancers. With many other scientists, we took part in a large scientific consortium called the Pan-Cancer Analysis of Whole Genomes (PCAWG), which studied a cohort of whole cancer genomes from over 2500 patients, the vast majority of which were adults. PCAWG published a suite of papers last year, including a study from my group on SSVs. SSV analysis requires whole-genome DNA sequencing. There have been studies of SSV-mediated gene deregulation in specific pediatric brain tumor types as well, including medulloblastoma. The CBTN brain tumor cohort that we examined was unique in that it had so many different brain tumor types represented that had not been studied in this way before.
Until recently, most genomic studies of human cancer have used "whole-exome" sequencing instead of whole-genome sequencing. Whole-exome sequencing involves looking for mutation in the coding regions of genes, focusing on the DNA components that make protein. Protein-coding genes make up about 1% of the human genome. Scientists used to consider the rest of the genome, the non-coding genome to represent "junk DNA", i.e., DNA that served no purpose. Now, we understand that the non-coding DNA is critical to proper regulation of the coding genes. In cancer, where non-coding DNA is damaged or altered, it can have profound effects on the over-expression of nearby genes.
The data and samples used for this study were drawn from the CBTN’s Pediatric Brain Tumor Atlas (PBTA), which is really noteworthy. While a lot of research at the CBTN happens amongst research teams across our member sites, your team hasn’t previously been a part of the CBTN’s collective efforts. How did you come to see the PBTA as a resource that could be useful for your study, and how were you able to use the data it holds?
The CBTN has made their genomic datasets available to the research community at large. Dozens of research projects using these data are ongoing, many of which are described at the CBTN.org website. With 854 tumors having combined whole-genome and gene expression data, we saw that the CBTN data represented a unique resource, where we could apply the analysis tools that we had previously developed in the study of adult cancers. The CBTN was very helpful in making the data available to my group, and the CBTN has hosted regular "office hours", where scientists like me can call in and ask any questions that we may have regarding the data.
What is the scope of this project? How many childhood cancers were you able to study, and what are some of the more noteworthy insights in the biology of these diseases that your team made?
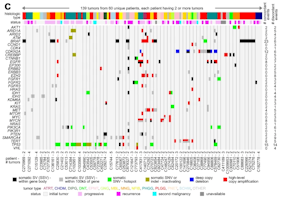
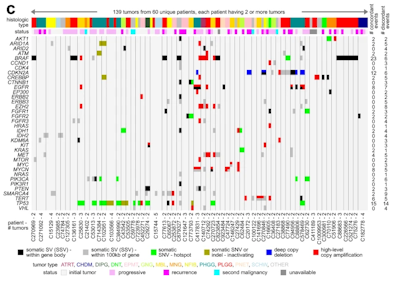
Using whole-genome and RNA sequencing from 854 tumors of more than 30 different types from the CBTN, we found that hundreds of genes had deregulated expression in association with the presence of nearby SSV breakpoints. SSVs extended the numbers of patients with tumors somatically altered for critical pathways, including receptor tyrosine kinases (KRAS, MET, EGFR, NF1), the Rb pathway (CDK4), the TERT oncogene, members of the MYC oncogene family (MYC, MYCN, MYB), and the HIPPO pathway (NF2). Compared to initial tumors, progressive or recurrent tumors involved a different set of SSV-gene associations. Tumors that had a lot of genomic rearrangements and SSVs in general also tended to have TP53 mutations, histone H3.3 gene H3F3C mutations, and high expression of DNA damage response genes.
Where do we go from here? How do you think the discoveries that came from this research might influence treatment for childhood brain cancer patients?
SSVs represent a previously under-appreciated mechanism by which cancer-promoting genes are altered. Knowing the set of genes targeted by DNA alteration in different cancers may lead to new possibilities for targeting specific genes using therapeutics. Different oncogenes would also converge on specific downstream pathways, and being able to infer what those pathways are could offer possibilities on ways to treat a larger subset of patients. Much of what is known today of personalized or precision medicine would focus on alterations within coding DNA, but SSVs represent alterations outside of coding DNA that would not be represented in current precision medicine approaches but could be included later.
related
Resources
Project

Data
Ongoing
Defining the Global Impact of Somatic Structural Variation on the Transcriptome of Human Pediatric Brain Cancers
Somatic structural variants (SSVs) are mutations arising in tumors that involve large segments of DNA. Using data from the Pediatric Brain Tumor Atlas, researchers will identify SSV patterns in pediatric cancer types that could lead to new therapies.
All Brain Tumor Types

Chad Creighton
Publication