Aggressive triple negative breast cancers have unique molecular signature on the basis of mitochondrial genetic and functional defects
Abstract
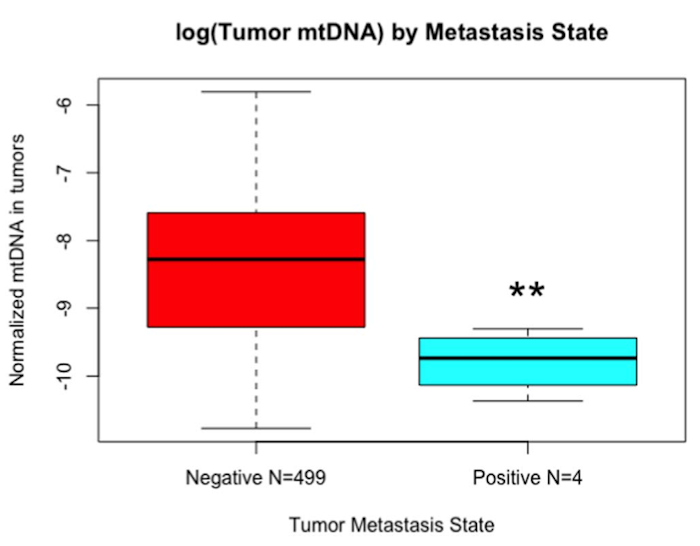
Metastatic breast cancer is a leading cause of cancer-related deaths in women worldwide. Patients with triple negative breast cancer (TNBCs), a highly aggressive tumor subtype, have a particularly poor prognosis. Multiple reports demonstrate that altered content of the multicopy mitochondrial genome (mtDNA) in primary breast tumors correlates with poor prognosis. We earlier reported that mtDNA copy number reduction in breast cancer cell lines induces an epithelial-mesenchymal transition associated with metastasis. However, it is unknown whether the breast tumor subtypes (TNBC, Luminal and HER2+) differ in the nature and amount of mitochondrial defects and if mitochondrial defects can be used as a marker to identify tumors at risk for metastasis. By analyzing human primary tumors, cell lines and the TCGA dataset, we demonstrate a high degree of variability in mitochondrial defects among the tumor subtypes and TNBCs, in particular, exhibit higher frequency of mitochondrial defects, including reduced mtDNA content, mtDNA sequence imbalance (mtRNR1:ND4), impaired mitochondrial respiration and metabolic switch to glycolysis which is associated with tumorigenicity. We identified that genes involved in maintenance of mitochondrial structural and functional integrity are differentially expressed in TNBCs compared to non-TNBC tumors. Furthermore, we identified a subset of TNBC tumors that contain lower expression of epithelial splicing regulatory protein (ESRP)-1, typical of metastasizing cells. The overall impact of our findings reported here is that mitochondrial heterogeneity among TNBCs can be used to identify TNBC patients at risk of metastasis and the altered metabolism and metabolic genes can be targeted to improve chemotherapeutic response.