Developing Cancer Informatics Applications and Tools Using the NCI Genomic Data Commons API
Abstract
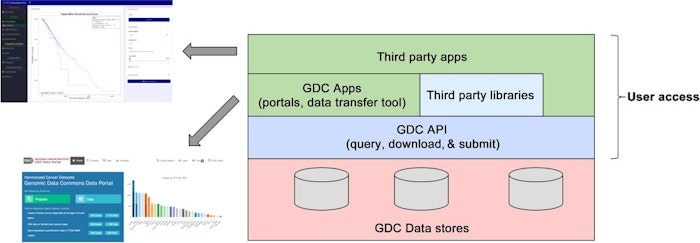
The NCI Genomic Data Commons (GDC) was launched in 2016 and makes available over 4 petabytes (PB) of cancer genomic and associated clinical data to the research community. This dataset continues to grow and currently includes over 14,500 patients. The GDC is an example of a biomedical data commons, which collocates biomedical data with storage and computing infrastructure and commonly used web services, software applications, and tools to create a secure, interoperable, and extensible resource for researchers. The GDC is (i) a data repository for downloading data that have been submitted to it, and also a system that (ii) applies a common set of bioinformatics pipelines to submitted data; (iii) reanalyzes existing data when new pipelines are developed; and (iv) allows users to build their own applications and systems that interoperate with the GDC using the GDC Application Programming Interface (API). We describe the GDC API and how it has been used both by the GDC itself and by third parties.