Histone Variant and Cell Context Determine H3K27M Reprogramming of the Enhancer Landscape and Oncogenic State
Abstract
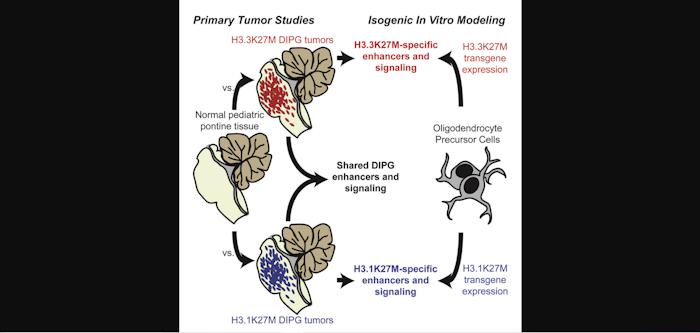
Development of effective targeted cancer therapies is fundamentally limited by our molecular understanding of disease pathogenesis. Diffuse intrinsic pontine glioma (DIPG) is a fatal malignancy of the childhood pons characterized by a unique substitution to methionine in histone H3 at lysine 27 (H3K27M) that results in globally altered epigenetic marks and oncogenic transcription. Through primary DIPG tumor characterization and isogenic oncohistone expression, we show that the same H3K27M mutation displays distinct modes of oncogenic reprogramming and establishes distinct enhancer architecture depending upon both the variant of histone H3 and the cell context in which the mutation occurs. Compared with non-malignant pediatric pontine tissue, we identify and functionally validate both shared and variant-specific pathophysiology. Altogether, we provide a powerful resource of epigenomic data in 25 primary DIPG samples and 5 rare normal pediatric pontine tissue samples, revealing clinically relevant functional distinctions previously unidentified in DIPG.
Highlights -
H3.3K27M and H3.1K27M DIPG tumors exhibit distinct active enhancer profiles
Extensive chromatin studies in primary H3K27M samples reveal oncogenic signaling
Disparate genomic locations of each H3K27M variant disrupt different PRC2 targets
H3K27M remodeling differs between NPCs and OPCs