Prognostic Implications of Immune-related Eight-gene Signature in Pediatric Brain Tumors
Abstract
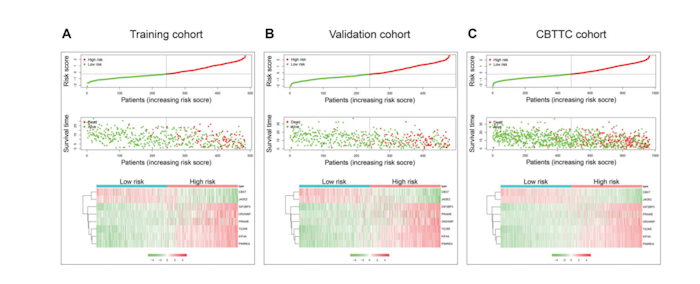
Genomic studies have provided insights into molecular subgroups and oncogenic drivers of pediatric brain tumors (PBT) that may lead to novel therapeutic strategies. Participants of the cohort Pediatric Brain Tumor Atlas: CBTTC (CBTTC cohort), were randomly divided into training and validation cohorts. In the training cohort, Kaplan-Meier analysis and univariate Cox regression model were applied to preliminary screening of prognostic genes. The LASSO Cox regression model was implemented to build a multi-gene signature, which was then validated in the validation and CBTTC cohorts through KaplanMeier, Cox, and receiver operating characteristic curve (ROC) analyses. Also, gene set enrichment analysis (GSEA) and immune infiltrating analyses were conducted to understand function annotation and the role of the signature in the tumor microenvironment. An eight-gene signature was built, which was examined by Kaplan-Meier analysis, revealing that a significant overall survival difference was seen, either in the training or validation cohorts. The eight-gene signature was further proven to be independent of other clinic-pathologic parameters via the Cox regression analyses. Moreover, ROC analysis demonstrated that this signature owned a better predictive power of PBT prognosis. Furthermore, GSEA and immune infiltrating analyses showed that the signature had close interactions with immune-related pathways and was closely related to CD8 T cells and monocytes in the tumor environment. Identifying the eight-gene signature (CBX7, JADE2, IGF2BP3, OR2W6P, PRAME, TICRR, KIF4A, and PIMREG) could accurately identify patients’ prognosis and the signature had close interactions with the immunodominant tumor environment, which may provide insight into personalized prognosis prediction and new therapies for PBT patients.
Acknowledgements
Chao Ma and Huan Luo thank Zhengzhou University Overseas Virtual Research Institute. Chao Ma thanks China Scholarship Council (No. 201708410121). We acknowledge support from the German Research Foundation (DFG) and the Open Access Publication Fund of Charité - Universitätsmedizin Berlin.