Landscape of Germline Histone Mutations in Pediatric Brain Tumor Patients
Email Principal Investigator
About this
Project
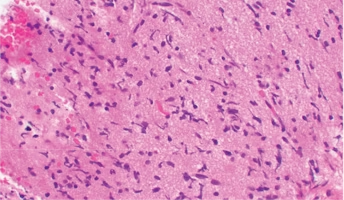
The majority of diffuse midline glioma (DMG) cases harbor mutations affecting histone H3-encoding genes. Histones are the proteins that pack DNA together, allowing them to have epigenetic (or non-gene) influences on tumors. The remaining 20% of DMG cases are H3WT DMGs, and the underlying genetic cause of H3WT DMGs remains unclear. Emerging data indicates an association between histone mutations that are passed down and developmental/neurological disorders and recent studies have begun to identify such mutations. However, there has yet to be extensive research into the landscape of genetic alterations affecting histone-encoding genes in DMG. Researchers propose to investigate germline mutations affecting histone-encoding genes in pediatric central nervous system tumors, and in particular diffuse midline gliomas (DMGs), from data provided by the Children’s Brain Tumor Network. The objective of this project is to analyze the inheritable (germline) genomes of DMG and other pediatric CNS tumors in the hopes of improving diagnostics, prognostics, and therapeutic options. This work is made possible through access to comprehensive data in the Pediatric Brain Tumor Atlas.
Ask The
Scientists
What are the goals of this project?
Researchers will investigate mutations affecting histone-encoding genes in pediatric central nervous system tumors, and in particular diffuse midline gliomas (DMGs), in the pursuit of identifying new driver histone variants to improve overall outcome for patients.
What is the impact of this project?
A deeper understanding of DMG types and their genetic drivers will in turn lead to advancements in the understanding the development and care of such tumors.
Why is the CBTN request important to this project?
This work would not be possible without the provision of high quality tumor data through access to the Pediatric Brain Tumor Atlas.
Meet The
Team
Erin Bonner
Institutions

Primary
Children’s National Hospital
Joined onEach year, the Brain Tumor Institute at Children’s National evaluates more than 100 new patients with brain tumors, and is recognized as a world leader in childhood brain tumor care and research. Children’s National has pioneered novel pediatric brain tumor therapies, including new molecularly-targe

University of Oslo
Center for Molecular Medicine Norway (NCMM)