Comparison of Fusion Calling Platforms in Pediatric DIPG and High‐grade Glioma
Email Principal Investigator

Carl Koschmann
CBTN Specimen
Backer
Internal funding
About this
Project
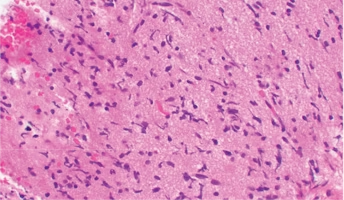
Molecular research into pediatric brain tumors is stimulating the development and testing of targeted therapies. However, effective targeted options are still unavailable for diffuse intrinsic pontine glioma (DIPG). DIPGs are especially lethal due to their aggressiveness and a lack of therapeutic options. In previous studies, researchers have found that 20% of pediatric high grade gliomas (pHGG) have gene fusions, genes formed of two previously independent genes. These fusions can frequently be pivotal in the growth and persistence of a tumor. Identification of gene fusions associated with pediatric DIPG could open the door for new precision medicine techniques. The collaborative team of researchers on this project include a bioinformatics group that specializes in the optimization of the analysis necessary to locate fusion genes. This analysis, the CODAC pipeline, has advanced research of precision medicine for adult solid tumors. In this project, researchers will compare CODAC to multiple other tools on DIPG data made available through the Pediatric Brain Tumor Atlas. Researchers are optimistic this will lead to an optimized platform for the identification of novel targets for the treatment of DIPG.
Ask The
Scientists
What are the goals of this project?
Researchers main goal of this project is to utilize a new analysis to identify new leads in the development of targeted therapies for patients with DIPG.
What is the impact of this project?
Similar analysis has been pivotal in defining new therapies for adult patients and this project will potentially expand the analysis’s impact to help pediatric patients with DIPG.
Why is the CBTN request important to this project?
The data available through the Pediatric Brain Tumor Atlas will allow researchers to expand this successful work to the pediatric population.
Specimen Data
The Children's Brain Tumor Network contributed to this project by providing access to the Pediatric Brain Tumor Atlas.
Meet The
Team
Marcin Cieslik, bioinformatician