Comprehensive Genomic and Immune Signature Profiling of Ependymoma and Diffuse Intrinsic Pontine Glioma
Email Principal Investigator

Kathleen M. Schieffer
CBTN Data
Backer
Internal funding
About this
Project
Brain tumors are the most common form of pediatric tumor, and while advancements have been made in treating a variety of tumor types, the need for comprehensive research into all pediatric brain tumors is great. Certain subtypes of ependymoma are associated with poor outcomes for patients as well as with diffuse intrinsic pontine glioma (DIPG) brain tumor patients. Both would benefit greatly from an expansion of comprehensive study of their genome. This study aims to do just that by taking an approach that combines genetic and epigenetic (non-gene) influences with data on immune response. The Pediatric Brain Tumor Atlas will give researchers access to a unique set of data necessary to carry out this research.
Ask The
Scientists
What are the goals of this project?
Researchers will perform comprehensive analysis on ependymoma and DIPG, including a comparison and contrast of their genetic underpinnings.
What is the impact of this project?
Ependymoma and DIPG are associated with poor outcomes for patients. Comprehensive understanding of the genetic, epigenetic, and immune response of these tumors will pave the way for the development of more effective treatments.
Why is the CBTN request important to this project?
The broad data available through the Pediatric Brain including rare specimens of ependymoma and DIPG is necessary to complete this comprehensive analysis.
Specimen Data
The Children's Brain Tumor Network contributed to this project by providing access to the Pediatric Brain Tumor Atlas.
Meet The
Team

700 Children's Drive Columbus, OH 43205


262 Danny Thomas Place, Memphis, TN 38105

Student Academic Services Building, 281 W. Lane Avenue, Columbus, OH 43210

Institutions

Nationwide Children's Hospital


St. Jude Children's Research Hospital
St. Jude is leading the way the world understands, treats and defeats childhood cancer and other life-threatening diseases. The mission of St. Jude Children’s Research Hospital is to advance cures, and means of prevention, for pediatric catastrophic diseases through research and treatment. Consisten


The Ohio State University
related
Histologies
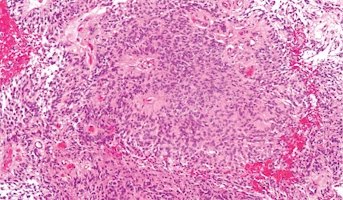
Ependymoma
Ependymomas arise from ependymal cells that line the ventricles and passageways in the brain and the center of the spinal cord. Ependymal cells produce cerebrospinal fluid (CSF). These tumors are classified as supratentorial or infratentorial. In children, most ependymomas are infratentorial tumors
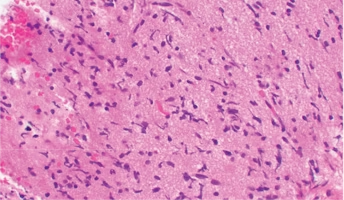
Diffuse Intrinsic Pontine Glioma
A presumptive diagnosis of DIPG based on classic imaging features, in the absence of a histologic diagnosis, has been routinely employed. Increasingly however, histologic confirmation is obtained for both entry into research studies and molecular characterization of the tumor.[5] New approaches with

