Discovering Novel Epigenetic Dependencies in Pediatric High-grade Glioma
Email Principal Investigator

Mariella Filbin
CBTN Specimen
CBTN Participants
CBTN Pre-clinical Models
About this
Project
Primary resistance to chemotherapy is a major challenge in treating brain tumors today. Finding novel and specific tumor dependencies is therefore essential for effective drug development and improving patient outcomes.
High-grade gliomas (HGG) are the most aggressive brain tumors in children and young adults. They are uniformly fatal despite multimodal treatment strategies combining surgery, radiation and chemotherapy. High-grade gliomas of the brainstem, termed diffuse intrinsic pontine glioma (DIPG), have an especially grim prognosis - median survival has not changed over the last few decades and is currently still less than 10 months after diagnosis.1 As a first breakthrough in the field, deep sequencing efforts in 2012 revealed unique and highly specific K27M mutations in histone 3.3 and 3.1 side chains in more than 90% of DIPG and midline glioma cases2-4, whereas G34R/V mutations were exclusively found in histone 3.3 of hemispheric high-grade gliomas in about 40% of cases. However, the potential therapeutic applications of this discovery have not yet been systematically studied.
I hypothesize that we can exploit these epigenetic mutations to develop new therapeutic approaches. Specifically, I hypothesize that (I) the abovementioned histone tail mutations generate an aberrant epigenetic landscape that renders DIPG and HGG cells hypersensitive to specific epigenetic perturbations, thus offering an opportunity to screen epigenetic regulators for identifying unique tumor vulnerabilities.
Ask The
Scientists
What are the goals of this project?
The goal of this project is to test the prediction that disabling the stem cell-like chromatin state of pediatric HGGs will induce cell death and differentiation.
What is the impact of this project?
Little progress has been made in the management of pediatric DIPGs and HGGs, even in the post-genomic era. New technologies are now enabling insight into their biology at a depth that was unthinkable even a few years ago: the CRISPR/Cas9 system allows investigation into the role of hundreds of epigenetic modifiers in DIPG/HGG in a single pooled screen. Applying these new technologies to precious patient derived tumor samples will shed unprecedented light on unique tumor vulnerabilities and dependencies that are not identifiable by genetic studies alone. The proposed research will provide an unparalleled view of epigenetically-mediated networks underlying HGG biology, and moreover, reveal novel tumor vulnerabilities that could ultimately lead to a cure for this deadly disease.
Specimen Data
The Children's Brain Tumor Network contributed to this project by providing cell lines.
related
Histologies
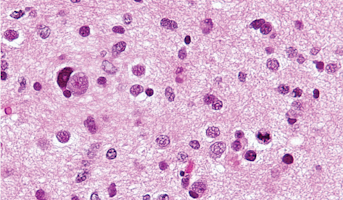
High-Grade Glioma
High-grade Gliomas (HGG) or astrocytomas in children nearly always result in a dismal prognosis. Although novel therapeutic approaches are currently in development, preclinical testing has been limited, due to a lack of pediatric-specific HGG preclinical models. These models are needed to help test
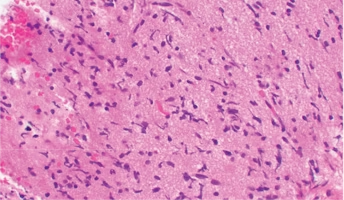
Diffuse Intrinsic Pontine Glioma
A presumptive diagnosis of DIPG based on classic imaging features, in the absence of a histologic diagnosis, has been routinely employed. Increasingly however, histologic confirmation is obtained for both entry into research studies and molecular characterization of the tumor.[5] New approaches with

