Proteogenomic Identification of Structural Variations
Email Principal Investigator

Brian Rood
CBTN Data
CBTN Specimen
CBTN Participants
CBTN Samples
Backer
Philanthropic Foundation Support
About this
Project
Cancers commonly arise as the result of changes affecting the DNA sequence of cells. These changes include small variations such as insertions and deletions as well as structural variations (SVs) including deletions, duplications, and translocations. In addition to these, tumor specific alterations can also give rise to proteins not commonly found in normal tissues. Those abnormal proteins are formed when SVs cause two different genes to form a new gene called gene fusions. Tumor specific fusion proteins could be potential targets for immunotherapy and DNA SVs represent potential biomarkers detectable by liquid biopsy. Researchers on this project seek to identify genomic alterations by detecting peptides, the building blocks of proteins, present in tumor cells. They will do so by assessing RNA sequencing data, analysis that reveals the presence of RNA in a biological sample, provided through the Pediatric Brain Tumor Atlas. Researchers have also been provided with rare, high quality tissue and RNA samples by the Children’s Brain Tumor Network. The integration of protein-based and genomic data provides a more comprehensive view of the biological features that drive cancer. Using this approach, identified proteins can be studied individually to clarify their role in the origin and growth of tumors and leveraged for use in therapeutics.
Ask The
Scientists
What are the goals of this project?
The main goal of this project is to assess RNA sequencing data to understand the presence of tumor specific fusion proteins that could serve as biomarkers or therapy targets.
What is the impact of this project?
The process researchers will use in this project allows for fusion proteins to be studied individually for this usefulness as biomarkers or targets for immunotherapy, advancing the field of personalized therapeutics for brain cancer.
Why is the CBTN request important to this project?
This project requires quality specimens and comprehensive data on pediatric brain cancers, all of which was provided by the Children’s Brain Tumor Network as tissue samples and access to the Pediatric Brain Tumor Atlas.
Specimen Data
The Children's Brain Tumor Network provided flash frozen tissue samples for proteomic peptide sequencing, CSF for ddPCR and RNA samples for targeted long read nanopore sequencing. CBTN also provided access to the Pediatric Brain Tumor Atlas.
Meet The
Team

Washington, DC, USA

Washington, DC, USA

Washington, DC, USA
related
Histologies
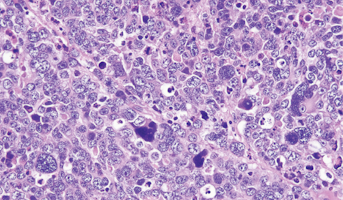
Medulloblastoma
Medulloblastomas comprises the vast majority of pediatric embryonal tumors and by definition arise in the posterior fossa, where they constitute approximately 40% of all posterior fossa tumors. Other forms of embryonal tumors each make up 2% or less of all childhood brain tumors.The clinical feature
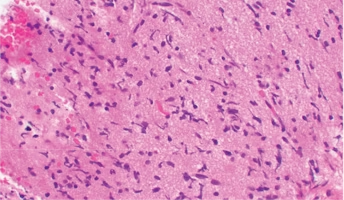
Diffuse Intrinsic Pontine Glioma
A presumptive diagnosis of DIPG based on classic imaging features, in the absence of a histologic diagnosis, has been routinely employed. Increasingly however, histologic confirmation is obtained for both entry into research studies and molecular characterization of the tumor.[5] New approaches with

