Analysis of Chromatin Pathways as Regulators of High Grade Glioma Gene Expression Patterns
Email Principal Investigator
About this
Project
Patients with high grade gliomas (HGG) face considerable treatment challenges and comprehensive research is needed to find new therapeutic options. Using the Pediatric Brain Tumor Atlas, researchers will identify genes that are expressed differently in patient-derived tumor samples compared to normal tissue samples. Gene expression, the process by which the genetic information is used to direct production of proteins in the body, is crucial in understanding the similarities and differences across tumor types. Additionally, these genetic signatures will be used in comparison to lab-grown tumor cell lines that have been genetically manipulated or treated with various drugs. Researchers hope to analyze these comparisons and identify patterns in gene expression that could pave the way for the development of new therapies.
Ask The
Scientists
What are the goals of this project?
Researchers will identify and compare tumor samples, normal tissue samples, and lab grown tumor samples in an effort to identify patterns in gene expression.
What is the impact of this project?
The identification of patterns in gene expression across tumor types could lead to the development of new targeted therapies.
Why is the CBTN request important to this project?
The breadth and depth of data available through the Pediatric Brain Tumor Atlas will be pivotal in supporting this project.
Specimen Data
The Children's Brain Tumor Network contributed to this project by providing access to the Pediatric Brain Tumor Atlas.
Meet The
Team

Boston, MA USA

Boston, MA

Boston, MA

Boston, MA


1125 Trenton Harbourton Rd, Titusville, NJ 08560
Cambridge, MA, USA

Institutions

Boston Children's Hospital

Harvard Medical School
Harvard Medical School is the graduate medical school of Harvard University and is located in the Longwood Medical Area of Boston, Massachusetts. Harvard Medical School has affiliation agreements with 15 of the world’s most prestigious hospitals and research institutes, vital partners that provide c

The Janssen Pharmaceutical Companies of Johnson and Johnson


Broad Institute of MIT and Harvard
related
Histologies
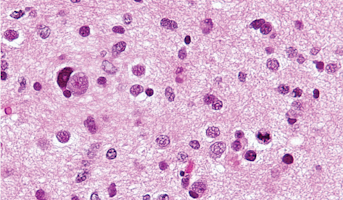
High-Grade Glioma
High-grade Gliomas (HGG) or astrocytomas in children nearly always result in a dismal prognosis. Although novel therapeutic approaches are currently in development, preclinical testing has been limited, due to a lack of pediatric-specific HGG preclinical models. These models are needed to help test
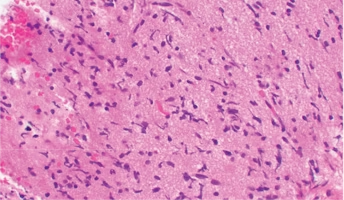
Diffuse Intrinsic Pontine Glioma
A presumptive diagnosis of DIPG based on classic imaging features, in the absence of a histologic diagnosis, has been routinely employed. Increasingly however, histologic confirmation is obtained for both entry into research studies and molecular characterization of the tumor.[5] New approaches with
related


