Identifying the Cells of Origins of Brain Tumors Using Somatic Mutations
Email Principal Investigator

Gilad Evrony
CBTN Data
CBTN Specimen
Backer
National Institute of Health
Sontag Foundation
About this
Project
Primary brain tumors are remarkably diverse, with more than 100 different defined types. The large diversity of brain tumors approximates the diversity of cell types in the normal brain, suggesting that each type of brain tumor corresponds to a different cell of origin. Definitive evidence that an individual brain tumor or type of tumor arises from a particular cell type requires a research method that allows for the tracing of single cells. Prior work by researchers on this project has developed methods for sequencing the entire genomes of single cells from the brain. This has revealed remarkable levels of mutations that could be used to trace the origins of brain tumors directly in human tissues. The goals of this project are to apply novel single-cell genome sequencing technologies for lineage tracing of pediatric brain tumors and to analyze genomic data generated by the Children’s Brain Tumor Network from pediatric tumors to profile mutational processes. The CBTN has already sequenced many pediatric brain tumors, which can immediately be used by researchers. Comprehensive genomic sequencing of these tumor types is not available anywhere else.
Ask The
Scientists
What are the goals of this project?
Researchers will apply genome sequencing technologies to pediatric brain tumor data in an effort to identify their cellular origin.
What is the impact of this project?
Understanding the cellular origin of various tumor types could lead to new therapeutic avenues.
Why is the CBTN request important to this project?
The comprehensive genomic data provided by the Pediatric Brain Tumor Atlas allows researchers to begin their analysis quickly and efficiently.
Specimen Data
The Children's Brain Tumor Network contributed to this project by providing access to the Pediatric Brain Tumor Atlas and by providing samples from post-mortem collections. (Gift from a Child)
Meet The
Team
related
Histologies
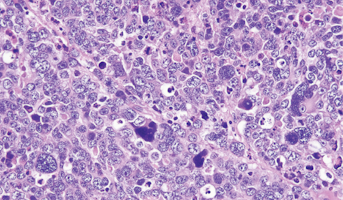
Medulloblastoma
Medulloblastomas comprises the vast majority of pediatric embryonal tumors and by definition arise in the posterior fossa, where they constitute approximately 40% of all posterior fossa tumors. Other forms of embryonal tumors each make up 2% or less of all childhood brain tumors.The clinical feature
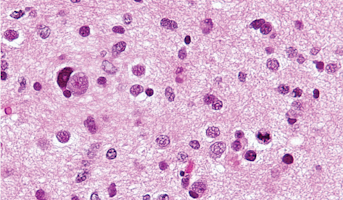
High-Grade Glioma
High-grade Gliomas (HGG) or astrocytomas in children nearly always result in a dismal prognosis. Although novel therapeutic approaches are currently in development, preclinical testing has been limited, due to a lack of pediatric-specific HGG preclinical models. These models are needed to help test
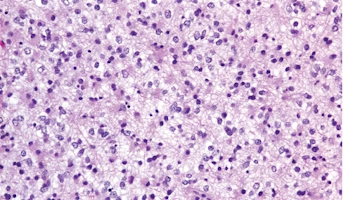
Low-Grade Glioma
Low-Grade Gliomas also called astrocytomas are the most common cancer of the central nervous system in children. They represent a heterogeneous group of tumors that can be discovered anywhere within the brain or spinal cord. Although surgical resection may be curative, up to 20% of children still su




