Pediatric Brain Tumor miRNA Profiling for the Cohort of Children’s Brain Tumor Network Specimens
Email Principal Investigator

Mateusz Koptyra
CBTN Specimen
CBTN Participants
CBTN Samples
Backer
Institutional Funds
About this
Project
The leading cause of disease related death in children is pediatric brain tumors. Surgery remains the main form of treatment for such tumors, however many brain tumors are unable to be fully removed with surgery. There are many factors that could lead to new and more effective therapies, including greater understanding of tumor genetics, comprehensive preclinical tests, and sensitive methods to assess tumor response to treatment. Mapping tumor molecular characteristics towards the discovery of tumor-related biomarkers, or observable changes in the body related cancer progression could lead to advancements in therapies, preclinical tests, and ongoing assessments. Using RNA samples provided by CBTN, researchers will produce a significant amount of biomarker data to be used in pediatric cancer research around the world.
Ask The
Scientists
What are the goals of this project?
The CBTN samples for this project have genomic data generated and associated with each, researchers seek to add RNA sequencing data for all of these samples in an effort to further characterize the tumors and to drive biomarker discovery.
What is the impact of this project?
Mapping tumor molecular characteristics and discovery and analysis of tumor-related biomarkers could lead to advancements in therapies, preclinical tests, and ongoing assessments.
Why is the CBTN request important to this project?
The RNA samples provided by CBTN will be integral to the data collection and analysis of this project.
Specimen Data
The Children's Brain Tumor Network contributed to this project by providing RNA samples for the generation of miRNAseq.
Meet The
Team
related
Histologies
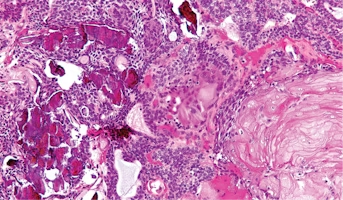
Craniopharyngioma
Childhood craniopharyngiomas are rare tumors usually found near the pituitary gland (a pea-sized organ at the bottom of the brain that controls other glands) and the hypothalamus (a small cone-shaped organ connected to the pituitary gland by nerves).Craniopharyngiomas are usually part solid mass and
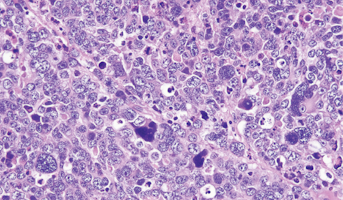
Medulloblastoma
Medulloblastomas comprises the vast majority of pediatric embryonal tumors and by definition arise in the posterior fossa, where they constitute approximately 40% of all posterior fossa tumors. Other forms of embryonal tumors each make up 2% or less of all childhood brain tumors.The clinical feature
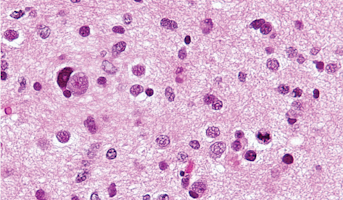
High-Grade Glioma
High-grade Gliomas (HGG) or astrocytomas in children nearly always result in a dismal prognosis. Although novel therapeutic approaches are currently in development, preclinical testing has been limited, due to a lack of pediatric-specific HGG preclinical models. These models are needed to help test
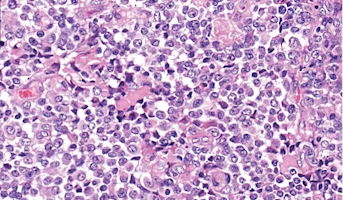
Atypical Teratoid/Rhabdoid Tumor
Central nervous system (CNS) atypical teratoid/rhabdoid tumor (AT/RT) is a very rare, fast-growing tumor of the brain and spinal cord. It usually occurs in children aged three years and younger, although it can occur in older children and adults. About half of these tumors form in the cerebellum or
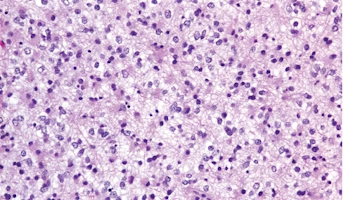
Low-Grade Glioma
Low-Grade Gliomas also called astrocytomas are the most common cancer of the central nervous system in children. They represent a heterogeneous group of tumors that can be discovered anywhere within the brain or spinal cord. Although surgical resection may be curative, up to 20% of children still su
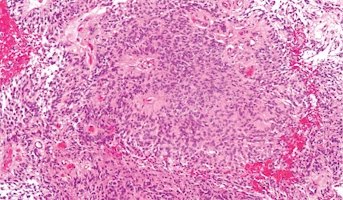
Ependymoma
Ependymomas arise from ependymal cells that line the ventricles and passageways in the brain and the center of the spinal cord. Ependymal cells produce cerebrospinal fluid (CSF). These tumors are classified as supratentorial or infratentorial. In children, most ependymomas are infratentorial tumors
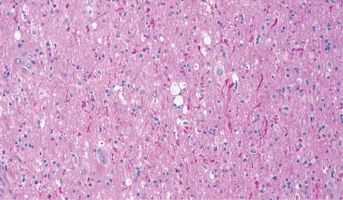
Ganglioglioma
Ganglioglioma presents during childhood and into adulthood. It most commonly arises in the cerebral cortex and is associated with seizures, but also presents in other sites, including the spinal cord.[65,74]The unifying theme for the molecular pathogenesis of ganglioglioma is genomic alterations lea
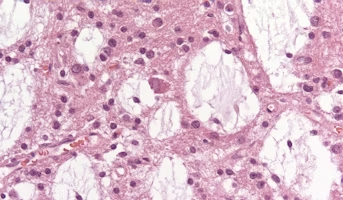
Dysembryoplastic Neuroepithelial Tumor
Septal DNET generally presents with symptoms related to obstructive hydrocephalus.[69,70] Septal DNET has an indolent clinical behavior, with most tumors not requiring treatment other than surgery. In a single-institution series that incorporated other literature-reported cases, the median age at pr
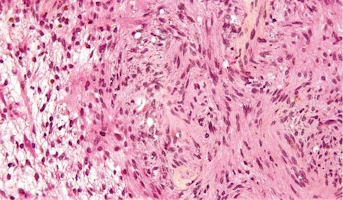
Schwannoma
Schwannoma is a rare type of tumor that forms in the nervous system. Schwannoma grows from cells called Schwann cells. Schwann cells protect and support the nerve cells of the nervous system. Schwannoma tumors are often benign, which means they are not cancer. But, in rare cases, they can become can





