Children's Brain Tumor Network Pediatric Brain Tumor Proteomics Pilot
Email Principal Investigator

Adam Resnick
CBTN Data
CBTN Specimen
CBTN Participants
CBTN Samples
About this
Project
Pediatric brain tumors are the leading cause of disease related death in children. Major factors contributing to treatment failures for children with brain tumors include: i) the lack of comprehensive molecular description of the disease and an associated dearth of integration of the tumors’ biological signatures at gene, protein and RNA levels; ii) paucity of comprehensive and molecularly-driven preclinical tests for informing precision-targeting disease pathways; and iii) lack of sensitive methods of assessing tumor response to treatment. Furthermore, while surgery remains the mainstay of pediatric brain tumor treatment, many tumors are non-resectable and/or progressive/disseminated tumors lead to significant tumor-related morbidity. In contrast to adults, tumors in central nervous system are often challenging to treat with traditional chemotherapy and radiation resulting in long-term neuro-toxicities in the context of the developing brain.
While pediatric cancers have benefited from large-scale genomics efforts lead by the NIH in the context of TARGET (Therapeutically Applicable Research To Generate Effective Treatments), brain tumors are not represented in these cohorts. The consortia-based efforts of the Children's Brain Tumor Network and the Pacific Pediatric Neuro-Oncology Consortium and associated platforms were complemented with the launch of CAVATICA (cavatica.org), a scalable, cloud-based storage and compute environment providing comprehensive, secure, and integrated access to pediatric cancer genomic data that is further integrated with additional rare-disease-genomic data. These early efforts are now undergoing significant expansion through the Commons Fund’s Kids First Program and CHOP’s role in leading the development of the Kids First Data Resource Center (DRC) along with its partners at the Ontario Institute for Cancer Research, the University of Chicago, Children’s National Health System, the Oregon Health and Science University and Seven Bridges. The Kids First DRC will support accelerated scientific discovery across more than >20,000 WGS from dozens of childhood cancer and structural birth defects cohorts being sequenced over the next 2 years. It is in this context of emerging genomic data and a data commons architecture that we propose to define a discovery cohort for comprehensive proteomic analysis in pediatric brain tumors.
Ask The
Scientists
What are the goals of this project?
We propose to perform comprehensive quantitative and associated post translational analyses on a pilot cohort
What is the impact of this project?
Our own and other’s efforts in characterizing pediatric brain tumors have defined their biologic underpinnings as distinct from adult brain tumors. Compared to adults, pediatric cancers possess significantly fewer driver mutations. Furthermore, in addition to canonical respective loss/gain of tumor suppressors/oncogenes, gene fusions events provide a common driving genetic mechanism. However, such structural variations also impinge on the epigenetic landscape in ways only recently appreciated. Indeed, especially in high-grade tumors, epigenetic dysregulation provides a common and ubiquitous paradigm for pediatric brain tumors likely informed by the developmental context of the central nervous system. Our own recent proteomic analyses of DIPGs, where histone mutations (“onco-histones”) have been identified, have focused on histone posttranslational modification and the potential therapeutic role of HDACs in modulating such epigenetic dysregulation. To date, few proteomic studies have been defined for pediatric cancer.
Why is the CBTN request important to this project?
The CBTN collects longitudinal, deep clinical, and phenotypic data on all prospective patients. More than 900 distinct flash-frozen specimens have been collected since 2011 across all brain tumor histologies with >75 mg of tissue and paired blood (germline) and are undergoing WGS/RNAseq (~200 samples have been completed).
Specimen Data
The Children's Brain Tumor Network contributed to this project by contributing over 200 samples and by providing access to the Pediatric Brain Tumor Atlas.
Meet The
Team
● Fred Hutch: Amanda Paulovich
● Harvard: Steven Gygi
● NIH: Kinsinger, Christopher (NIH/NCI)
● NIH: Rodriguez, Henry (NIH/NCI)
● NIH/NCI: Boja, Emily (NIH/NCI)
● NIH/NCI: Bocik, William (NIH/NCI)

Philadelphia, PA, USA

Washington, DC, USA

Washington, DC, USA






NCI Shady Grove Campus, 9609 Medical Center Drive, Rockville, MD 20850
Institutions

Primary
Operations Center

Children’s Hospital of Philadelphia
Joined onOperations Center for the Children’s Brain Tumor Tissue Consortium, the Children’s Hospital of Philadelphia (CHOP) is currently ranked 1st nationally for their Pediatric Cancer Program by U.S. News & World Report. CHOP’s Biobank is home to the CBTTC’s pediatric brain and CNS tumor biorepository; the

Primary
Children’s National Hospital
Joined onEach year, the Brain Tumor Institute at Children’s National evaluates more than 100 new patients with brain tumors, and is recognized as a world leader in childhood brain tumor care and research. Children’s National has pioneered novel pediatric brain tumor therapies, including new molecularly-targe


National Cancer Institute, National Institute of Health
related
Histologies
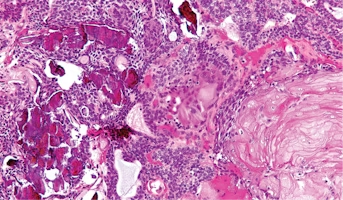
Craniopharyngioma
Childhood craniopharyngiomas are rare tumors usually found near the pituitary gland (a pea-sized organ at the bottom of the brain that controls other glands) and the hypothalamus (a small cone-shaped organ connected to the pituitary gland by nerves).Craniopharyngiomas are usually part solid mass and
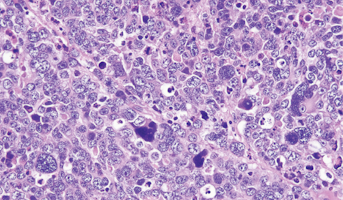
Medulloblastoma
Medulloblastomas comprises the vast majority of pediatric embryonal tumors and by definition arise in the posterior fossa, where they constitute approximately 40% of all posterior fossa tumors. Other forms of embryonal tumors each make up 2% or less of all childhood brain tumors.The clinical feature
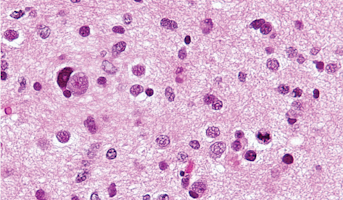
High-Grade Glioma
High-grade Gliomas (HGG) or astrocytomas in children nearly always result in a dismal prognosis. Although novel therapeutic approaches are currently in development, preclinical testing has been limited, due to a lack of pediatric-specific HGG preclinical models. These models are needed to help test
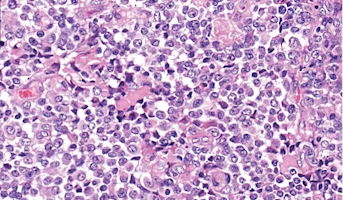
Atypical Teratoid/Rhabdoid Tumor
Central nervous system (CNS) atypical teratoid/rhabdoid tumor (AT/RT) is a very rare, fast-growing tumor of the brain and spinal cord. It usually occurs in children aged three years and younger, although it can occur in older children and adults. About half of these tumors form in the cerebellum or
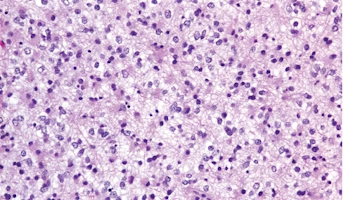
Low-Grade Glioma
Low-Grade Gliomas also called astrocytomas are the most common cancer of the central nervous system in children. They represent a heterogeneous group of tumors that can be discovered anywhere within the brain or spinal cord. Although surgical resection may be curative, up to 20% of children still su
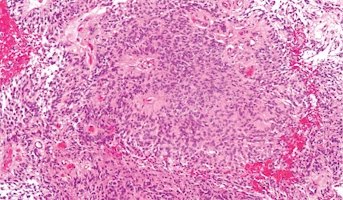
Ependymoma
Ependymomas arise from ependymal cells that line the ventricles and passageways in the brain and the center of the spinal cord. Ependymal cells produce cerebrospinal fluid (CSF). These tumors are classified as supratentorial or infratentorial. In children, most ependymomas are infratentorial tumors
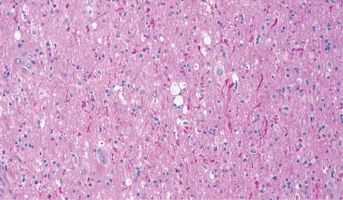
Ganglioglioma
Ganglioglioma presents during childhood and into adulthood. It most commonly arises in the cerebral cortex and is associated with seizures, but also presents in other sites, including the spinal cord.[65,74]The unifying theme for the molecular pathogenesis of ganglioglioma is genomic alterations lea
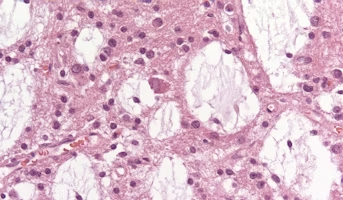
Dysembryoplastic Neuroepithelial Tumor
Septal DNET generally presents with symptoms related to obstructive hydrocephalus.[69,70] Septal DNET has an indolent clinical behavior, with most tumors not requiring treatment other than surgery. In a single-institution series that incorporated other literature-reported cases, the median age at pr
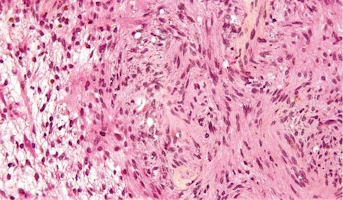
Schwannoma
Schwannoma is a rare type of tumor that forms in the nervous system. Schwannoma grows from cells called Schwann cells. Schwann cells protect and support the nerve cells of the nervous system. Schwannoma tumors are often benign, which means they are not cancer. But, in rare cases, they can become can
related
Resources
Project

Data
Specimen
Planning
Proteomic Analysis of CBTN Cell Lines
Using cell lines provided by the Children’s Brain Tumor Network, researchers will attempt to build resources used to develop effective treatments and predict the treatment outcomes of pediatric brain tumor patients.
HGG, (AT/RT)

Roger Reddel
Project

Specimen
Ongoing
Pediatric Brain Tumor miRNA Profiling for the Cohort of Children’s Brain Tumor Network Specimens
Many pediatric brain tumors are unable to be removed in surgery, requiring the development of new effective therapies. Using RNA samples provided by CBTN, researchers are producing important characterization of the tumors and biomarker data to be used by researchers around the world in the pursuit of such new approaches.
Craniopharyngioma, Medulloblastoma, HGG, (AT/RT), LGG, Ependymoma, Ganglioglioma, DNET, Schwannoma

Mateusz Koptyra
Project

Specimen
Ongoing
Project Hope: High-Grade Glioma-Omics in Pediatric and AYA
High grade gliomas can affect patients of all ages, and cross-age group analysis could lead to advancements in care for all populations. Using samples and data provided by the Children’s Brain Tumor Atlas, researchers supported by the NCI seek to improve outcomes for patients experiencing these tumors.
HGG

Adam Resnick

